Introduction
I have been in IT for almost 15 years and my primary focus is to collaborate with the customer on their goals and bring their IT strategy to the next level of digital transformation. I have been supporting customers in various verticals in the role as an Integrator, Presales Solutions Engineer and Solution Architect.
Today the world is changing rapidly with processes, application development and integration moving even faster. Workflows and their applications are highly automated and connect directly to private or shared data endpoints over the globe. Data growth, the distribution of data and connecting apps with the right Service Level Agreements (SLA) for availability, reliability and performance requirements is also a challenge in context with this rapid change and digital transformation.
In this blog post and next blog posts I will give you an overview about use cases of various industry branches where a next generation application as service and distributed storage is more important than ever before.
For all use cases I prefer to use an open source software centric approach which is independent from any hardware or cloud provider and driven by a huge community with over 15 million developers.
I will start this blog with a use case of the health care sector.
Use Case: Health Care & Next Generation Sequencing (NGS)
The health system is more strained and challenged than ever before. Researchers are fighting against diseases that did not exist before or only special analyses can detect. More than 300 million people worldwide live with a rare disease. In the search for a diagnosis, especially genome sequencing provides definitive answers to patients and their doctors.
In the context of growth, NGS architecture must be as flexible as possible, because new sequencing methods means that the throughput of processing data increases fivefold annually and the data growth doubles in the process. Nowadays, not only partial areas, but also a whole genome can be viewed. Around 4 years ago less than 1% of genetic information was decoded, where Next Generation Sequencing now achieves 3-5%.
The complete workflow is about patient-related data processing. In order to be able to carry this out in accordance with the law, the GDPR and other data privacy and other laws must be observed. For example, the Genetic Diagnostics Act sets the requirements for genetic testing of human beings and the act on the Violation of Private Secrets (German Criminal Code §203) ensures that practices are not allowed to use the public cloud. That can be changed in the future with additional regulations and it’s necessary to be prepared with the right solution which allows to build a hybrid cloud.
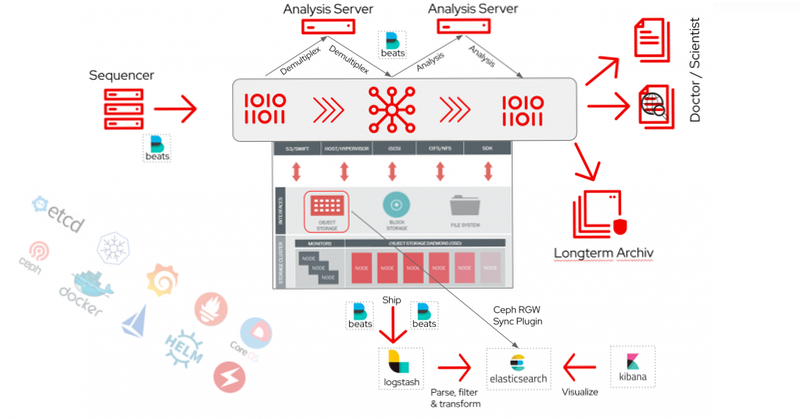
How does the process look like from patient blood sample to patient findings?

The sequencer performs an optical analysis of the sample, which is mapped through many cycles, and generates enormous amounts of data. These are demultiplexed to know which sequences originate from which samples. An analysis is then carried out and the results are available for evaluation.
These steps are carried out via a data pipeline and an appropriate processing and storage system in the backend, where the bioinformatician is primarily working on optimizing workflows and long-term archiving with automation by Ansible, own scripts and various tools like awscli, s3cmd, python scripts or native S3 interface in the application. Ansible also supports standardising the infrastructure and checks the environment for changes as required for security. The findings are now interpreted by the scientists and doctors and discussed with the patient to address possible therapies.
Customer success story: CLIMB
Red Hat Ceph Storage provides object storage and block storage with incredibly high performance,” said Connor. “We can use it to store copies of images on our system, so that other researchers can download and use them. The features of Red Hat Ceph Storage let us share these large volumes efficiently. We’re confident that we have the storage volume we need to keep up with future generations of research data.
Technical insights
The requirements for applications, services and storage include open and scalable APIs, as well as export functions of the audit logs to document all processes in accordance with the law and make them available for auditing and WORM functionalities to ensure immutability and upgradeability in a fast and secure manner.
The Institute for Medical Genetics and Applied Genomics at Tübingen University Hospital developed and distributed an application within an open source project with the name megSAP. It’s a NGS data analysis pipeline for medical genetics which runs tools for diagnostics, extensive logging, testing and makes sure that the results are valid. The strategic change from closed source to open source strengthened the project immensely.
( https://github.com/imgag/megSAP )
To install the tools from megSAP, it’s necessary to install some prerequisites packages upfront for RHEL8 which again builds the foundation for all applications and services with support for 24/7 customer needs:
Add LANGUAGE and LC_ALL to /etc/locale.conf for suppress perl warnings and fulfil installation dependencies:
LANGUAGE="en_US.UTF-8"
LANG="en_US.UTF-8"
LC_ALL="en_US.UTF-8"Code language: JavaScript (javascript)Install packages and dependencies:
# subscription-manager repos --enable codeready-builder-for-rhel-8-x86_64-rpms
# yum groupinstall "Development Tools"
# yum install zlib-devel bzip2-devel xz-devel ncurses-devel libcurl-devel cpan cpanminus gd-devel libdb-devel -y
# dnf install php-cli php-xml php-mysqlnd R-core R-core-devel -yCode language: PHP (php)Note: The default package version of php in RHEL8.3 is 7.2, but you can check it with following command # dnf module list php
Installing perl modules by “dnf install ‘perl(Config::General)’ -y” or cpanm is not necessary, because the installation script will do these tasks for you.
After the installation of the required tools the installation script should be ready to run successfully. Just following the “Initial setup” steps: https://github.com/imgag/megSAP/blob/master/doc/install_unix.md
Example of an installation output:
~/tools # circos-0.69-9/bin/circos -modulesCode language: JavaScript (javascript)
All required packages were installed into Red Hat Enterprise Linux 8 (minor release 3) for a NGS pipeline.
How do you fulfil your SLAs inside or outside your company?
Let’s now focus on this part where services are provided through the IT Services and Infrastructure team. OpenShift Container Platform runs containers and virtual machines and data can be easily stored via OpenShift Container Storage and Ceph for simplified management by an operator or plugin for a future-proof architecture. It allows you also to work with the storage via S3 (Simple Storage Service) bucket claim and attach data pools via buckets directly to the applications. That’s unique for a kubernetes management framework and not one else instead of Red Hat.
Just think about that the patient information has to be accessible by the scientist or doctor all the time with the right inhouse or external SLAs in a secure manner.

OpenShift is essentially an enterprise-ready open-source stack kubernetes / k8s orchestrator which gives you world-class enterprise support and services, deep expertise to confidently run any application in VM, container and serverless environments and continuous security.

The bioinformatician works on the workflow and how to handle a lot of data and information about the unstructured data, which reflects the “data exchange platform & security”. The findings from the patient must be guaranteed as quickly and securely as possible and there is also no downtime possible, because the processing runs mostly several days for high sensitive analysis.
The patient and sample information will be enriched by metadata during the workflow in order to be able to find and assign them quickly. The IT Services and Infrastructure provides processing server, databases, web server, search engines with an web-ui. Fast search engines are important to make unstructured content searchable – i.e. by elasticsearch and kibana – and create workflows to provide information via dynamic websites, backup newest results and archiving of long term content.
How do you connect your application in OpenShift to the data storage and work with them?
The underlying storage backend is OpenShift Container Storage as integrated single software defined storage platform to provide object, block and file out of the box with simplified UI and easy administration by IT-Administrator. It’s built on open source community projects like the Operator Framework, Rook and Ceph. On this foundation it supports multi-cloud and hybrid cloud deployments and allows storage management by policies.
With OpenShift Storage it’s easy to use object storage. OpenShift lets you provision buckets to the user or application really fast – just in 2 steps – or automated via api-driven workflows.



Get the Access Key and Secret Key with the right ACLs to allow access only to this bucket.
Additional information on following page: https://access.redhat.com/documentation/en-us/red_hat_openshift_container_storage/4.5/html-single/managing_openshift_container_storage/index#creating-an-object-bucket-claim-using-the-openshift-web-console_rhocs

However, a data pipeline is only useful if you distribute data between company locations or over the globe and access and write from various operating systems and application, i.e. Linux <-> Windows <-> macOS <-> Application-X.
OpenShift Container Platform as orchestrator for kubernetes / k8s with additional services like operators, Single-Sign On and security hardening helps to operate the heterogeneous environment of the NGS data pipeline. An operator for OpenShift Container Storage (OCS) provides a seamless integration into containers and virtual machines via object (S3 / Swift), file and block to fulfil the requirements for a fast data exchange between each other. In addition to that it simplified the management and provided a scalable backend to grow and connect with any cloud. OCS can be deployed directly in OpenShift worker, outside on separate commodity compute nodes or connected via operator with an external Ceph installation.
Tools
There are a wide range of tools to work with an S3 object storage via put/ingest, get/outgest, simulate file systems via FUSE-based file system backend or using metadata to the object self for search engines and event-driven workflows, such as
- s3cmd
- awscli
- s3fs
- Goofyfs
- Rclone
- Cloudberry Drive, S3Express (only Microsoft Windows)
- Native by python code (Boto3)
s3cmd
At first I want to show you s3cmd which can be downloaded from http://s3tools.org/s3cmd
It’s a command line S3 client for Linux and macOS and allows to manage data in Amazon AWS S3 and other S3 compatible storage providers, such as Red Hat Ceph Storage (Object Gateway daemon – radosgw).
The simplest way to configure is s3cmd –configure or directly by modifying the configuration file ~/.s3cfg
# cat ~/.s3cfg
[default]
access_key = XYZ
secret_key = XYZ
host_base = <FQDN>
host_bucket = <FQDN>
check_ssl_certificate = False
check_ssl_hostname = False
use_http_expect = False
use_https = True
signature_v2 = True
signurl_use_https = True
[...]Code language: PHP (php)List current buckets where you can store your data without any limit except physical space:
# s3cmd -c ~/.s3cfg ls
2020-11-09 15:13 s3://mschindl-s3Code language: PHP (php)Put / upload files to a bucket:
# s3cmd -c ~/.s3cfg put <local folder>/code-is-open.png s3://mschindl-s3/Code language: HTML, XML (xml)
Modify the metadata of the object which was uploaded before:
# s3cmd info s3://mschindl-s3/code-is-open.pngCode language: PHP (php)
# s3cmd modify --add-header x-amz-meta-accessgroup:external s3://mschindl-s3/code-is-open.pngCode language: PHP (php)
Now you can see additional information was added to the object and can be used.
Pre-Sign an URL with time stamp to provide fast access to the data secure by HTTPS, traceable by user and with an retention time:
# s3cmd -c ~/.s3cfg signurl s3://mschindl-s3/code-is-open.png $(echo "`date +%s` + 3600 * 24 * 7" | bc)Code language: PHP (php)
Such links can be embedded in dynamic websites in example for the sharing of information with a limited time frame, or just for sharing data without retention

AWScli
is similar to s3cmd and can be downloaded via RHEL8
# dnf install awscliCode language: PHP (php)or from https://github.com/aws/aws-cli
and has the advantage to manage your AWS instances, improve ingest throughput by parallel transfer functionality and it is also supported by AWS with a large community.
Filesystem in Userspace
Some workloads expect a mountpoint to work properly and in this case s3fs, goofyfs or Cloudberry Drive are tools to provide a file system gateway to solve this challenge. As you can see in the screenshots below s3fs was integrated into macOS via # brew install s3fs and configured with following steps:
# echo <AccessKey>:<SecretKey> > ${HOME}/.passwd-s3fs
# chmod 400 /Users/mschindl/.passwd-s3fs
# mkdir ~/s3fs
# s3fs mschindl-s3 ~/s3fs -o passwd_file=${HOME}/.passwd-s3fs -o url=https://s3-openshift-storage.apps.ocp4.stormshift.coe.muc.redhat.com/ -o use_path_request_style
Note: add "-o dbglevel=info -f -o curldbg > s3fs.err 2>&1" for troubleshootingCode language: PHP (php)

After mounting the bucket to the user file system via FUSE. I opened the picture and also added Finder specific information like the green point to it. An additional object (s3://mschindl-s3/._code-is-open.png ) with the information about the green point was created and is not usable for other applications than macOS Finder, but increases the object count on the storage backend.
Today, a lot of applications support S3 natively and it’s easy to connect to the S3 endpoint and workflows can use multiple data interfaces with the same data as shown below.
Last but not least a connection between an object storage and a search engine like elasticsearch is best practise and helps to understand what is happening and necessary for searching in an endlessly scaling unstructured storage backend. It also helps to fulfil the auditing regulations.
Elasticsearch, Logstash and Kibana (ELK) are Open Source software that are combined to create a monitoring system that can be integrated with Ceph. The ELK stack is easily customizable which makes it easy to define custom events, visualizations and dashboards.
There are two ways to process and insert logs and events into elasticsearch. First, beats ships logs from compute and storage to logstash and logstash parse, filter and transform them in a structured way for elasticsearch. Second, the object storage sends bucket notifications directly to elasticsearch. After that kibana visualizes the metadata information from elasticsearch and provides search capabilities.

Conclusion
Next Generation Sequencing is a complex topic and more important than ever before.
In the past, customers with NGS workloads had to struggle with more and more data growth and the increase in processing speed, where also new applications are only delivered as containers.
To keep up, customers are porting applications more and more to container platforms such as Kubernetes and switching data management to S3 object storage in order to be able to work directly with the data and to keep the data pipeline lean and efficient.
The key to efficient process and workflow design was the collaboration between the core and supporting departments.
Core departments include, for example, clinical genetics, molecular genetics and development. The supporting departments include, for example, IT Services & Infrastructure, Marketing and Quality Management.
A procedure for process and workflow optimization between the departments provides for discussions, demo environments, tests, evaluation of the advantages and disadvantages of, for example, open source and commercial applications and the optimization steps.
IT Services & Infrastructure manages the environment with virtual machines (i.e. Linux and Windows) and containers (virtualized, bare metal or as managed kubernetes in the cloud) and migrates the data from the classic storage system using simple open source tools such as rclone, awscli or s3cmd in Linux and macOS and Cloudberry Drive or S3Browser in Windows. As a result these data were processed and added with metadata in order to work with them more quickly and directly. The integration in elastic search helped to make an unstructured, endlessly scalable storage system (object storage) searchable and empowered to control workflows based on this metadata.
Red Hat, the world’s leading provider of open source software solutions with enterprise support, helps to implement solutions stacks from middleware to infrastructure by providing a variety of services, consulting, and training.
Just leave a comment if you wish more information about this topic or the products mentioned in this blog. Any feedback or collaboration is highly appreciated! Our code is open …